New publication: Increasing phenotypic annotation improves the diagnostic rate of exome sequencing in a rare neuromuscular disorder
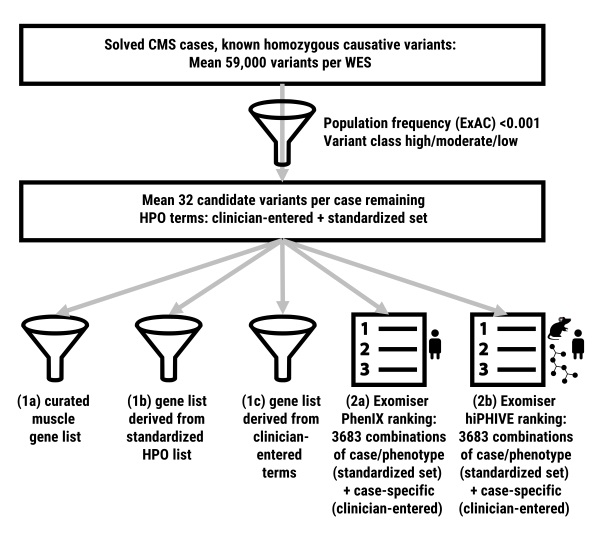
How can we use phenotypic descriptions to aid genetic diagnosis starting with hundreds of rare variants from next-generation sequencing results? How comprehensively should a doctor annotate a patient’s phenotype to help identify the causative variant? These questions are increasingly relevant for doctors seeing undiagnosed patients, but data showing how phenotypic annotation can be used to assist diagnosis in a real-world situation has until recently been lacking. In our new paper by Thompson et al published this week in Human Mutation, we reanalyse a set of patients already diagnosed with congenital myasthenic syndromes, showing how phenotype captured using the human phenotype ontology (HPO) can be integrated into gene-list filtering methods and how variant prioritisation tools such as Exomiser can be effective in homing in on the correct causative variant. In our restricted set of patients, gene-list filters created using HPO annotations performed similarly to the curated muscle gene table list, while Exomiser ranked the correct causative variant in first position in 90% of cases with seven HPO annotations compared to 62% based on variant data alone. Although we cannot automatically extrapolate these results to other disease areas, they provide a real-world validation of the utility of incorporating phenotype into diagnostic and discovery workflows.
Read an open-access version of the publication at the journal website here.
Thompson, R, Papakonstantinou Ntalis, A, Beltran, S, Töpf, A, de Paula Estephan, E, Polavarapu, K, ‘t Hoen PAC, Missier P, Lochmüller H.
Increasing phenotypic annotation improves the diagnostic rate of exome sequencing in a rare neuromuscular disorder.
Hum. Mutat. 2019.
PMID:31231902
Abstract

First author Rachel Thompson
Phenotype-based filtering and prioritization contribute to the interpretation of genetic variants detected in exome sequencing. However, it is currently unclear how extensive this phenotypic annotation should be. In this study, we compare methods for incorporating phenotype into the interpretation process and assess the extent to which phenotypic annotation aids prioritization of the correct variant. Using a cohort of 29 patients with congenital myasthenic syndromes with causative variants in known or newly discovered disease genes, exome data and the Human Phenotype Ontology (HPO)-coded phenotypic profiles, we show that gene-list filters created from phenotypic annotations perform similarly to curated disease-gene virtual panels. We use Exomiser, a prioritization tool incorporating phenotypic comparisons, to rank candidate variants while varying phenotypic annotation. Analyzing 3,712 combinations, we show that increasing phenotypic annotation improved prioritization of the causative variant, from 62% ranked first on variant alone to 90% with seven HPO annotations. We conclude that any HPO-based phenotypic annotation aids variant discovery and that annotation with over five terms is recommended in our context. Although focused on a constrained cohort, this provides real-world validation of the utility of phenotypic annotation for variant prioritization. Further research is needed to extend this concept to other diseases and more diverse cohorts.