New publication: Correction of pseudoexon splicing caused by a novel intronic dysferlin mutation
We were really pleased to collaborate with the Dominov and Brown labs at the University of Massachusetts Medical School. This research discovered a novel pathogenic point mutation in a dysferlin intron, thus determining the genetic cause in a group of patients with limb girdle muscular dystrophy. In addition, through performing targeted exon skipping we were able to correct the mutation in patient cells restoring the expression of normal dysferlin protein.
A patient involved in the study said “I am happy to see these results published because finally, after 47 years I have a definite diagnose, an explanation why the second mutation could not previously be identified, and have been given hope that a cure might be found in the future”.
Open access article is available to read here.
Abstract:
Objective: Dysferlin is a large transmembrane protein that functions in critical processes of membrane repair and vesicle fusion. Dysferlin-deficiency due to mutations in the dysferlin gene leads to muscular dystrophy (Miyoshi myopathy (MM), limb girdle muscular dystrophy type 2B (LGMD2B), distal myopathy with anterior tibial onset (DMAT)), typically with early adult onset. At least
416 pathogenic dysferlin mutations are known, but for approximately 17% of patients, one or both of their pathogenic variants remain undefined following standard exon sequencing methods that interrogate exons and nearby flanking intronic regions but not the majority of intronic regions. Methods: We sequenced RNA from myogenic cells to identify a novel dysferlin pathogenic
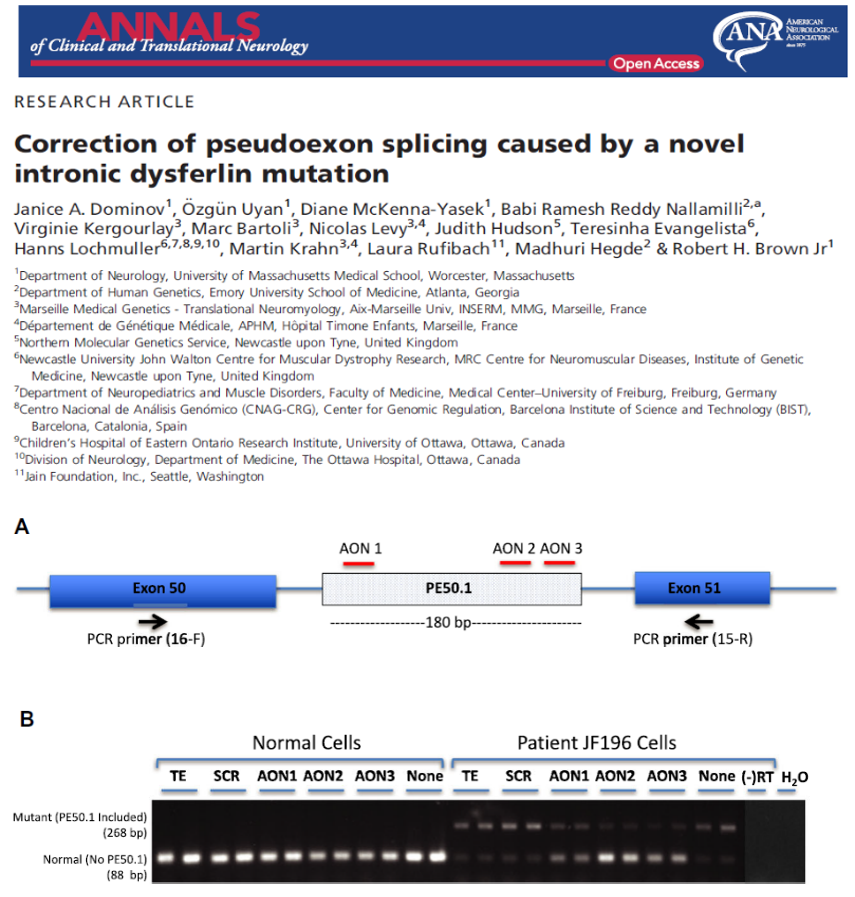
variant in two affected siblings that previously had only one disease-causing variant identified. We designed antisense oligonucleotides (AONs) to bypass the effects of this mutation on RNA splicing. Results: We identified a new pathogenic point mutation deep within dysferlin intron 50i. This intronic variant causes aberrant mRNA splicing and inclusion of an additional pseudoexon (PE, we term PE50.1) within the mature dysferlin mRNA. PE50.1 inclusion alters the protein sequence, causing premature translation termination. We identified this mutation in 23 dysferlinopathy patients (seventeen families), revealing it to be one of the more prevalent dysferlin mutations. We used AON-mediated exon skipping to correct the aberrant PE50.1 splicing events in vitro, which increased normal mRNA production and significantly restored dysferlin protein expression. Interpretation: Deep intronic mutations can be a common underlying cause of dysferlinopathy, and importantly, could be treatable with AONbased exon-skipping strategies.