Solve-RD flagship publication in Nature Medicine: Genomic reanalysis of a pan-European rare-disease resource yields new diagnoses
Patients and families with rare disorders often remain without a genetic diagnosis despite modern advances in diagnostic testing. Due to the rarity and global distribution of these diseases, data sharing at an international level is essential to make new discoveries and provide an end to the diagnostic odyssey faced by many individuals affected by an undiagnosed rare disorder. Genetic diagnosis requires the accurate identification and interpretation of genomic variants, which is particularly challenging in the rare disease field owing to lack of data, meaning that many variants captured by next-generation sequencing cannot correctly be interpreted as disease-causing at the time the sequencing is performed.
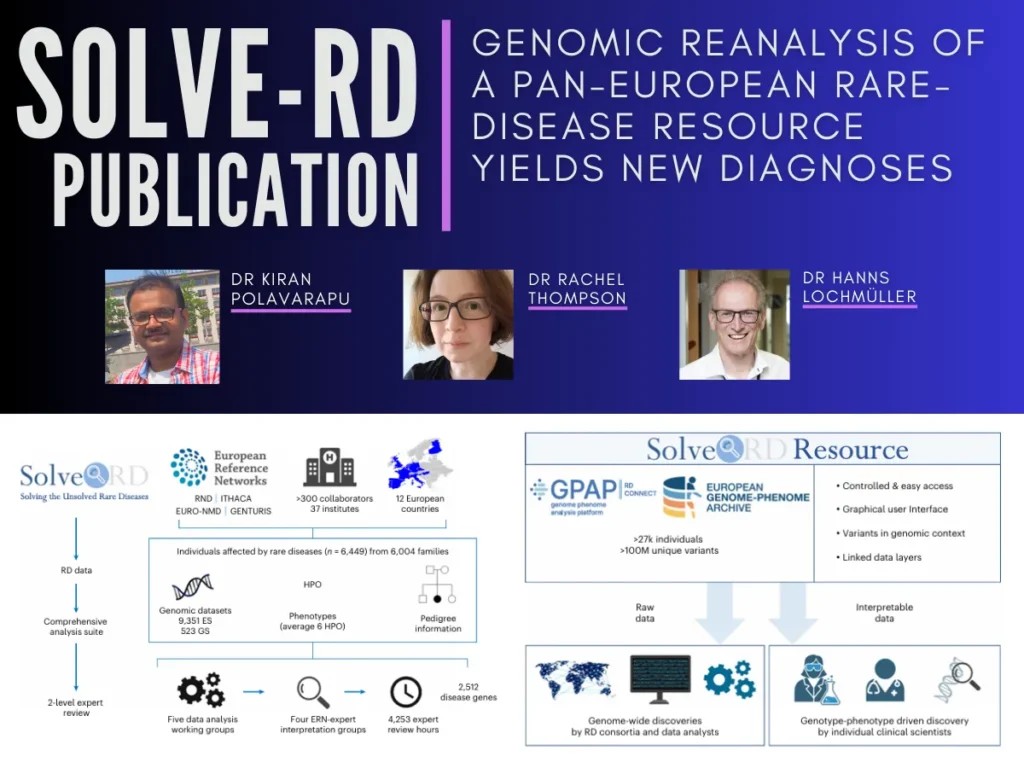
Our publication demonstrates the significant potential of reanalyzing existing genomic data, reinterpreting variants already captured by the sequencing experiment to establish their pathogenicity. It provides the results of a systematic reanalysis of data from 6,447 individuals with previously undiagnosed rare diseases from 6,004 families recruited from across Europe by the Solve-Rare Diseases Consortium (Solve-RD).
Authors and collaboration information
In this study, “Genomic reanalysis of a pan-European rare-disease resource yields new diagnoses”, published recently in the journal Nature Medicine, we worked as part of a major international consortium, Solve-RD, to develop a novel framework for genetic re-analysis of undiagnosed rare disorders, leading to the successful diagnosis and an end to the diagnostic odyssey for over 700 patients in whom a diagnosis had previously been elusive despite extensive testing. Lab postdoctoral researcher and CIHR fellowship recipient Dr Kiran Polavarapu is a joint first author on this major study together with young researchers from the other European Reference Networks (ERNs) who participated in the reanalysis and variant interpretation for their respective disease areas. The work was coordinated by teams from Barcelona, Spain and Nijmegen, The Netherlands. Steven Laurie from the Centro Nacional de Análisis Genómico (CNAG) led activities under the supervision of senior authors from the four participating ERNs together with Dr Sergi Beltran (CNAG) and Dr Alexander Hoischen (Radboud University, Nijmegen).
About the Study
Dr Kiran Polavarapu coordinated the reanalysis and variant interpretation of the neuromuscular disease (NMD) cohort within the study together with the senior author responsible for NMDs, Dr Ana Töpf. This cohort consisted of exome and genome datasets from 1517 undiagnosed families submitted to the Data Interpretation Task Force of ERN EURO-NMD. 260 of these unsolved neuromuscular families were submitted by the Lochmüller Lab, of which 32 (12.3%) obtained a diagnosis either through the systematic reanalysis or individual family-based analysis performed by Dr Polavarapu and the Lochmüller Lab drylab team under the supervision of Dr. Hanns Lochmüller using the RD-Connect Genome-Phenome Analysis Platform (GPAP).
Outcome
The results of the study demonstrate the significant potential of reanalyzing existing genomic data. The framework from this study, rare disease–reanalysis logistics (RD-REAL), with a two-level expert review, successfully provided a diagnosis for 506 families through structured reanalysis and 249 families through local expert review with an overall diagnostic rate of 12.6%. 84.1% of novel diagnoses came from newly interpretable variants, while 15.9% resulted from rare variant types previously undetectable by standard bioinformatic methods. Importantly, potential medical actionability was identified for 14.4% of cases, meaning that the result did not simply provide the patient with an end to the search for a diagnosis but also potentially enabled a change in medical care.
The study shows that even after a decade of diagnostic exome sequencing, new interpretations remain possible, and it is hoped that the infrastructure and collaborative networks set up by Solve-RD can serve as a blueprint for future further scalable international efforts. The resource is open to the global rare-disease community, allowing phenotype, variant and gene queries, as well as genome-wide discoveries.
Reference
Laurie, S., Steyaert, W., de Boer, E., Polavarapu, K., Schuermans, N., Sommer, A. K., Demidov, G., Ellwanger, K., Paramonov, I., Thomas, C., Aretz, S., Baets, J., Benetti, E., Bullich, G., Chinnery, P. F., Clayton-Smith, J., Cohen, E., Danis, D., de Sainte Agathe, J. M., Denommé-Pichon, A. S., … Hoischen, A. (2025). Genomic reanalysis of a pan-European rare-disease resource yields new diagnoses. Nature medicine, 31(2), 478–489. https://doi.org/10.1038/s41591-024-03420-w
Click here to access the manuscript!